It’s all about sequencing these days. Let’s sequence this, let’s sequence that. We have already sequenced the human genome (The Human Genome Project (HGP)), we have discovered an overwhelming biodiversity in the global ocean, there’s The Earth Microbiome Project (EMP) and NIH Human Microbiome Project. Scientific society is taking full advantage of the fact that the cost of sequencing has been consistently decreasing over the last few decades while the quality and speed of sequencing have been going in the opposite direction. Additionally, we have been getting better and better with bioinformatics post-sequencing analysis which allowed us to leverage modern day computational techniques to unravel secrets hidden in the genomic code. How did it all begin, though? Let’s have a brief look at the journey of sequencing that’s taken us from 1972—when Walter Fiers sequenced the DNA of a complete gene (the gene encoding the coat protein of the bacteriophage MS2) by digesting the viral RNA, isolating oligonucleotides, and separating them via electrophoresis/chromatography—to modern days when the entire HGP could be accomplished in less than two weeks and at a cost of just around $ 1,000 (with time and prices continuously dropping).
DNA sequencing timeline
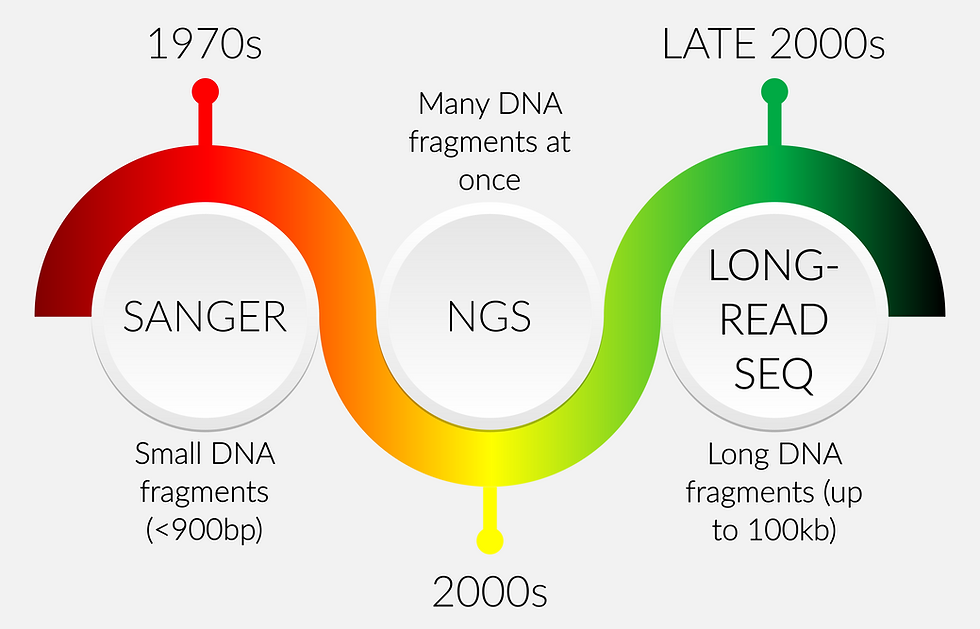
DNA sequencing timeline
Sanger
It all started with Sanger technology which was developed by Dr. Frederick Sanger in the 1970s and has been regarded as the gold standard for many years (until the appearance of NGS based technologies). This method is typically used to sequence relatively small DNA fragments (< 900 base pairs [bp]). The process is based on the detection of fluorescently labeled chain-terminating nucleotides (dideoxyribonucleotides or ddNTPs) that are incorporated by a DNA polymerase during the replication of a template. The resulting labeled DNA fragments are then mapped out to determine the original DNA sequence.
NGS
The next major step in the sequencing evolution occurred when next generation sequencing (NGS) took over the sequencing world in the 2000s. NGS is a massively parallel sequencing technology that is capable of sequencing many DNA fragments at once. NGS techniques are faster and cheaper compared to Sanger sequencing, and they allow more efficient determination of long DNA sequences.
Long-read DNA sequencing technology
Finally, the third-generation DNA sequencing or long-read DNA sequencing was first introduced in 2008–2009. This sequencing technology can read the DNA sequences that typically range between 10,000 and 100,000 base pairs (bp). Two main players in the current long-read sequencing marketplace are Oxford Nanopore sequencing and PacBio single-molecule real-time (SMRT) sequencing. Even though these two techniques are capable of sequencing long lengths of DNA, they are based on different methodologies which will be covered later in this article.
The third generation of DNA sequencing
Let’s have a closer look at this third generation sequencing. Is it better? Is it cheaper? Is it faster?
As mentioned earlier, currently two main long-read DNA sequencing technologies dominate the market: PacBio sequencing and Oxford Nanopore sequencing. These platforms apply different scientific principles in their core technology but they both can produce long DNA sequences.
PacBio
In 2010, PacBio introduced its zero-mode waveguide (ZMW) that uses “nanoholes” containing a single DNA polymerase. In this case, incorporation of a single uniquely labeled nucleotide can be observed directly. Different levels of a signal emitted during incorporation are then recorded by detectors attached below the ZMW.
Oxford Nanopore Technologies
On the other hand, Oxford Nanopore Technologies introduced the GridION in 2012 that measures changes in ionic current that occur when DNA fragments are moved through biological nanopores. Different DNA sequences produce different levels of electrical resistance while passing through these pores, thus allowing to determine the exact nucleotide sequence.
One of the clear advantages of the Nanopore technology is that sequencing devices based on it are very portable and can be used anywhere in the world. For example, MinION weighs only 100 g (~3.5 oz) and can be run in the field while connected to a laptop. Over the years, this technology has been used in many exciting projects including DNA sequencing of soil samples during an expedition on an icecap in Iceland, studying beetles living in remote caves in Montenegro, and studying crop infecting viruses on African farms.
Portable DNA sequencer
Advantages of long-read DNA sequencing technology
Long-read sequencing has several inherent advantages compared to short-read NGS technologies.
One of the main advantages is the ability of long-read sequencing to more accurately generate de novo genomic assemblies (when no reference genome is available). Since long-read sequencing is capable of spanning much longer regions, it allows for more accurate detection of structural variants (large insertions or deletions), regions with complex genetic rearrangements, and genomic repeats or copy number variations (CNV). Accurate CNV detection plays an important role in the diagnosis and prediction of many genetic diseases including developmental delay, schizophrenia, and autism. Another important advantage of long-read sequencing is the ability to identify long haplotypes – determination of which variant is on which copy of a chromosome. Given that DNA consists of two strands on each chromosome and both strands are sequenced at the same time, it is difficult to determine “what variant goes with what variant on the same strand” using short-read sequencing (SRS).
Alongside this impressive list of advantages, a few limitations of long-read sequencing still exist that prevent their wider spread at research and clinical institutions.
The accuracy per read is lower than in SRS (mostly due to the inability to control the speed with which the DNA molecules pass through the pore – these are systematic errors).
The cost per base is higher and overall sample throughput is lower than in SRS.
Downstream bioinformatics approaches are less developed, although this gap is being actively narrowed.
What’s next in the sequencing domain?
The general consensus for what the sequencing future holds is a play on the “more for less” slogan. Technological advances will most likely lead to even faster and more accurate sequencing methods. At the same time, future approaches are expected to require less input DNA which is especially important while working with difficult-to-get samples.
What’s probably even more important than the chemical side of the sequencing evolution is what’s going to happen in the data analysis domain. Further advances in bioinformatics, machine learning, and AI approaches applied to sequencing data will lead to more robust and accurate biological predictions and clinical diagnoses.
Whether we should expect more rapid evolution of short- or long-read sequencing techniques, or the combination of the two (or yet something new entirely) is something to look out for.
References:
Fiers, W., Contreras, R., Duerinck, F., Haegeman, G., Iserentant, D., Merregaert, J., Min Jou, W., Molemans, F., Raeymaekers, A., Van den Berghe, A. and Volckaert, G., 1976. Complete nucleotide sequence of bacteriophage MS2 RNA: primary and secondary structure of the replicase gene. Nature, 260(5551), pp.500-507. doi: 10.1038/260500a0.
Gowers, G.O.F., Vince, O., Charles, J.H., Klarenberg, I., Ellis, T. and Edwards, A., 2019. Entirely off-grid and solar-powered DNA sequencing of microbial communities during an ice cap traverse expedition. Genes, 10(11), p.902. doi: 10.3390/genes10110902
Maestri, S., Cosentino, E., Paterno, M., Freitag, H., Garces, J.M., Marcolungo, L., Alfano, M., Njunjić, I., Schilthuizen, M., Slik, F. and Menegon, M., 2019. A rapid and accurate MinION-based workflow for tracking species biodiversity in the field. Genes, 10(6), p.468. doi: 10.3390/genes10060468
Portable DNA sequencer helps farmers stymie devastating viruses. L Shaffer, 2019. https://www.pnas.org/doi/full/10.1073/pnas.1901806116
Comments